Introduction
Kidney transplantation (KT) remains the most common solid organ transplant, with over 25,000 procedures performed annually. Despite the increasing number of transplants, long-term graft survival has stagnated due to the lack of sensitive, non-invasive diagnostic methods. Kidneys from extended criteria donors (ECD) and donors after circulatory death (DCD) face higher risks of delayed graft function, injury, and rejection, necessitating improved monitoring techniques.
Proteomics has emerged as a promising tool for non-invasive KT evaluation by analyzing thousands of proteins in biological samples such as biopsies, blood, urine, and perfusion fluids. This approach allows early detection of graft damage, identification of transplant pathologies, and prediction of short- and long-term graft function. Proteomics-based diagnostics provide a multiplex, high-throughput, and highly sensitive alternative to traditional biomarker-based assessments, which are often limited in scope.
Recent advancements in mass spectrometry, AI-driven bioinformatics, and data processing platforms have enhanced the accuracy and efficiency of proteomic analysis. These technologies enable comprehensive protein profiling, improving diagnostic precision and transplant monitoring. As proteomics continues to evolve, it holds significant potential to revolutionize KT diagnostics, facilitating early intervention and better patient outcomes.
Materials and Methods
The review aimed to identify recent advances in proteomic diagnostics for kidney graft evaluation and explore correlations between identified proteins. A systematic review was conducted following PRISMA guidelines using PubMed and Scopus databases. Studies focused on proteomic biomarkers in kidney graft injury and function were included, with eligibility based on specific criteria such as research in human urine or blood samples and quantitative analysis methodologies.
Study Selection
A systematic selection process was followed, retrieving studies from PubMed and Scopus without publication date restrictions. Titles and abstracts were screened independently by two reviewers, and eligible articles were fully assessed. Studies focusing on proteomics in kidney graft evaluation were included, while reviews, meta-analyses, letters, and non-English publications were excluded. Data on study characteristics, diagnostic sensitivity, and protein biomarkers were extracted and categorized using the UniProt database.
Categorization of biomarkers based on gene ontology annotations
Proteomic biomarkers were classified into blood-based and urine-based categories, with further subdivisions for blood-based proteomics. The STRING database was used for gene set enrichment analysis (GSEA), classifying proteins based on biological processes, molecular functions, and cellular components. Significant signaling pathways linked to kidney dysfunction were retrieved from databases like REACTOME and WikiPathways. Statistical analysis determined key hub-proteins and their relevance to kidney graft evaluation.
Results
Study selection and characteristics of included studies
The study selection followed PRISMA 2020 guidelines. A total of 1,754 manuscripts were identified from PubMed and Scopus. After removing 159 duplicates, 87 studies met initial eligibility criteria. Following a detailed review, 23 studies were excluded for lacking proper quantitation techniques, and 47 did not focus on proteomics in relevant complications. Ultimately, 17 studies met all criteria and were included in this systematic review.
Sample and population characteristics The median number of samples was 59 (interquartile range 117), whereas 12 (70.58%) studies had under 100 samples. Out of 17 proteomics studies, 13 used samples derived from urine and four used blood-based samples. Three of the blood-based samples were plasma and one study employed serum samples. Figure 2 shows quantitative data on biological sample selection in all included samples.
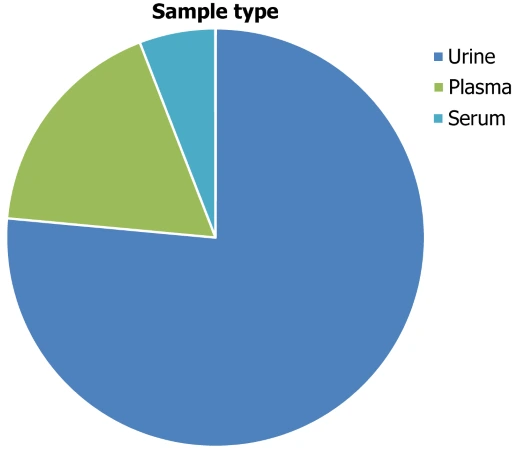
Figure 2. Quantitative data about the biological sample selection in all the included samples.
Evaluation techniques
The most used method in the reviewed studies was LC-MS/MS, either targeted (MRM) or untargeted (discovery). MRM provides higher sensitivity for specific analytes, while untargeted LC-MS/MS detects a broad range but may lack precision for individual proteins. Some studies combined both to improve accuracy.
ELISA was commonly used for validation, with direct and sandwich ELISA ensuring specificity. Other techniques included targeted urine proteome assay, isobaric tags for quantitation, serine hydrolase activity profiling, and proximity extension assay.
These methods collectively enhanced the identification and quantitation of biomarkers in kidney graft evaluation.
Biological samples proteome
In 17 studies, 58 proteomic biomarkers were analyzed. CXCL10 was the only overlapping protein. Of these, 23 were found in urine, 33 in plasma, and 2 in serum. Among them, 39 were elevated, 12 were downregulated, 6 indicated better prognosis, 1 was specific to injured kidney grafts, and 1 showed increased activity in complicated KT cases.
Protein categorization and enrichment analysis based on GO terms
GO enrichment analysis of 58 proteins from healthy and pathological samples identified key BP, CC, and MF terms. In complicated KT cases, proteins were linked to catalytic activity, immune response, and stress defense. CC analysis showed most proteins in the extracellular space, exosome, and matrix, with many being secreted. MF terms highlighted signaling receptor binding and molecular function regulation. For proteins associated with good outcomes, CC analysis found all in the extracellular space, with three in blood microparticles.
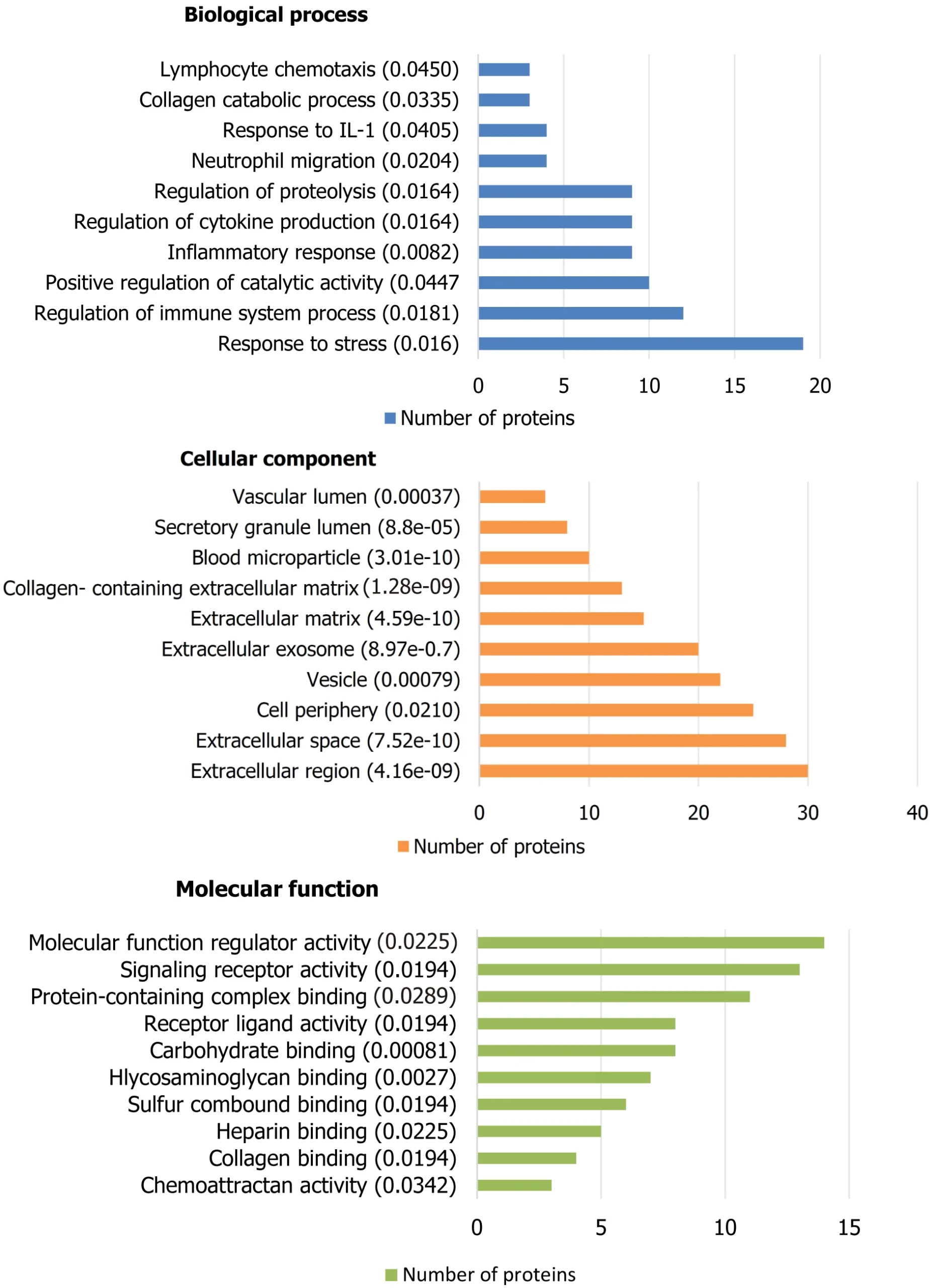
Figure 4. Gene ontology analysis of upregulated proteins detected on samples of kidney transplant recipients with graft injury or dysfunction. False discovery rate is mentioned in the parenthesis.
Application of proteomics in kidney graft evaluation
Proteomic biomarkers hold diagnostic and predictive value in kidney transplantation by correlating protein changes in biological fluids with biopsy findings and kidney function markers like creatinine and eGFR. Among 17 studies, five linked protein assessment to biopsy results, focusing on chronic allograft dysfunction, subclinical rejection, and fibrosis. Twelve studies explored proteomic biomarkers for graft function prediction, emphasizing specific time points post-transplantation. Several studies highlighted proteomics’ role in short- and long-term graft function assessment, with some demonstrating its superiority over traditional methods like creatinine evaluation.
Discussion
Renal transplantation is the most common treatment for end-stage kidney disease, offering better quality of life than dialysis. However, graft-related complications persist. Conventional monitoring relies on markers like serum creatinine and biopsies, which detect damage late. Proteomic analysis offers a promising non-invasive approach to predict complications early.
Key findings:
- Immune Response: Enrichment of IL-1 signaling suggests its potential as a therapeutic target.
- Fibrosis & Graft Injury: Proteins like Metalloproteinase-9 and Fibronectin are linked to fibrosis and long-term graft dysfunction.
- Amyloidosis & Coagulation: Some proteins correlate with amyloidosis and cardiovascular diseases, impacting graft health.
- Kidney Failure Markers: Proteins like Myoglobin and Angiotensin indicate graft damage.
- Protective Proteins: Prothrombin and Apolipoprotein A-I are linked to better transplant outcomes.
CXCL10, a key biomarker, is elevated in inflammatory kidney conditions. LC-MS/MS remains the preferred proteomic method, but data standardization challenges persist. Further research is needed for clinical application.
Conclusion
This systematic review showed that very few proteomic-level studies have been conducted to identify the key proteins associated with KT complications, and particularly with subclinical injury and prediction of graft function. Nevertheless, to the best of our knowledge, we were unable to locate any gene expression data regarding KT and non-invasive graft evaluation. We highlight the importance of new research studies to be held in order to identify the best possible panel of proteins for the early non/minimal invasive detection of different complications, as well as the best possible method of evaluation.
References
1. Thongprayoon C, Kaewput W, Pattharanitima P, Cheungpasitporn W. Progress and Recent Advances in Solid Organ Transplantation. J Clin Med. 2022;11.
2. Current Strategies for Living Donor Kidney Transplantation [Internet]. Hergiswil (CH): European Dialysis and Transplant Nurses Association/European Renal Care Association (EDTNA/ERCA); 2021–.
3. Alimi R, Hami M, Afzalaghaee M, Nazemian F, Mahmoodi M, Yaseri M, Zeraati H. Factors Affecting the Long-Term Survival of Kidney Transplantation in Northeastern of Iran between 2000 and 2015. Iran J Public Health. 2021;50:2076-2084.
4. Warmuzińska N, Łuczykowski K, Bojko B. A Review of Current and Emerging Trends in Donor Graft-Quality Assessment Techniques. J Clin Med. 2022;11:487.
5. Huang E, Mengel M, Clahsen-van Groningen MC, Jackson AM. Diagnostic Potential of Minimally Invasive Biomarkers: A Biopsy-centered Viewpoint From the Banff Minimally Invasive Diagnostics Working Group. Transplantation. 2023;107:45-52.
6. Psatha K, Kollipara L, Drakos E, Deligianni E, Brintakis K, Patsouris E, Sickmann A, Rassidakis GZ, Aivaliotis M. Interruption of p53-MDM2 Interaction by Nutlin-3a in Human Lymphoma Cell Models Initiates a Cell-Dependent Global Effect on Transcriptome and Proteome Level. Cancers (Basel). 2023;15:3903.
7. Ramalhete LM, Araújo R, Ferreira A, Calado CRC. Proteomics for Biomarker Discovery for Diagnosis and Prognosis of Kidney Transplantation Rejection. Proteomes. 2022;10.
8. Sirolli V, Piscitani L, Bonomini M. Biomarker-Development Proteomics in Kidney Transplantation: An Updated Review. Int J Mol Sci. 2023;24.
9. Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367-1372.
10. Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13:731-740.